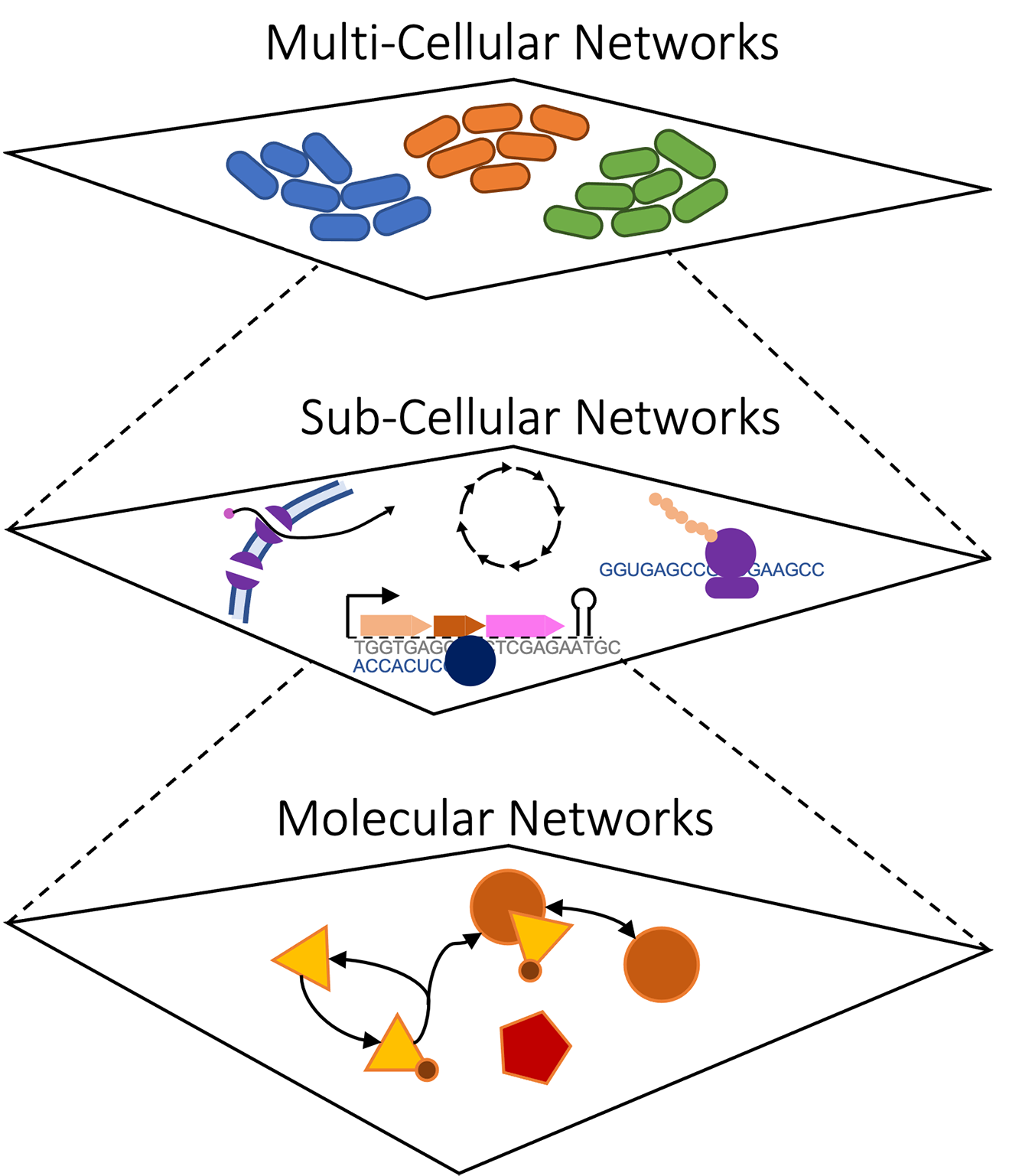
Our lab focuses on building comprehensive, multi-scale models of cells –- from their molecular underpinnings, to their integrated functions, to the population level with many heterogeneous cells interacting in a shared environment. Research in biology has generated abundant experimental data on the molecular composition, spatial organization, and dynamics of thousands of cell types; however, we need new tools that can transform this data into predictive models that can be used for scientific discovery. Rather than building isolated models that capture a narrow range of cellular behavior with a single class of representation, the next generation of computational models will need to combine *multi-source* and *multi-level* data with diverse, meaningful representations of biological mechanisms into integrative multi-scale simulations. These simulations will be used to interpret the input datasets, make experimental predictions, identify medical solutions, and help us understand fundamental principles of biological organization.
To support this vision, we are building Vivarium –- an open-source ecosystem of modular biological models. We have several projects that apply Vivarium in the domains of single cell bacterial physiology, community interactions in microbiomes, synthetic cells, and the origins of life.
Affiliations at UConn:
Center for Cell Analysis and Modeling
Department of Molecular Biology and Biophysics
Biomedical Engineering Department
Funding:
DARPA Simulating Microbial Systems
NIH Center for Reproducible Biomedical Modeling
NSF Center for Chemical Currencies of a Microbial Planet
DARPA Discovering Unknome Function
John Templeton Foundation Science of Purpose
About Vivarium:
vivarium-core
Vivarium Collective
Vivarium Documentation
Compositional Systems Biology and Process Bigraphs:
Foundations of a Compositional Systems Biology
Process Bigraph on Github
Process Bigraph Supplemental Materials
